|
|
| |
 |
MYOSIN
LIGHT CHAIN KINASES
|

|
 |
skMLCK
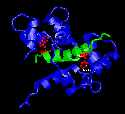
Figure 1
M13(skMLCK)
with Ca2+-Calmodulin
Created with RasMac v2.6
PDB ID=2BBM, 2BBN |
M13
peptide from skeletal muscle myosin light chain kinase
(skMLCK) was one of the first one to be studied structurally
with calmodulin. The structure was solved using the NMR
technique by Ikura et al
(1992). As predicted by many scientists before the
structure was solved, M13 peptide was indeed in an alpha-helix
conformation within calmodulin.
In Figure 1, two red residues are the two key bulky hydrophobic
residues (Trp4, Phe17) that serve to anchor the peptide
to calmodulin. The two bulky residues are separated by
12 residues in the between them. As one can see, the anchor
residues fit nicely into the binding pockets.
|
| It
was noted from early on that calmodulin has many metionine
residues. At first, the role of methionines were not known.
The structural study by Ikura et al (1992) revealed that
the methionine residues are in the binding pockets. Because
the side chains of methionine are very flexible, it was
predicted that many different bulky hydrophobic residues
can be accomodated in the binding pocket. |
smMLCK
|
| Shortly
after the NMR study was published, the X-ray crystalography
study on the smooth muscle myosin light chain kinase (smMLCK)
was completed by Meador
et al (1992). Although the method of structural analysis
was different, Meador et al showed that the smMLCK also
binds to calmodulin with two bulky hydrophobic residues
(Trp5, Leu18) serving as anchors. This at first suggested
that perhaps all calmodulin-binding peptides possess the
same properties. When the structure of CaMKII
with calmodulin was published (Meador
et al, 1993), 1-14 motif was no longer the sole binding
motif for calmodulin-binding peptides. Still, 1-14 motif
is the second largest motif in this database. |
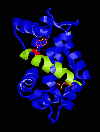
Figure 2
smMLCK
with Ca2+-Calmodulin
Created with RasMac v2.6
PDB ID = 1CDL
|
|
|
|
| |
| |
|
|
| |
|
|