|
|
| |
 |
CaM-DEPENDENT KINASE II
|

|
 |
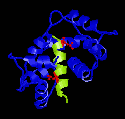
Figure 1
Peptide from CaMKII and Ca2+-Calmodulin
Created with RasMac v2.6
PDB ID = 1CDM, 1CM1 |
The
X-ray crystal structure of the peptide from the alpha
subunit of Calmodulin-dependent kinase II and calmodulin
was solved by Meader et
al in 1993. The structure is somewhat superficially
similar to those of skMLCK or smMLCK and Ca2+-Calmodulin.
Two bulky hydrophobic residues contact methionine residues
in the binding pocket of calmodulin. The overall conformation
of the complex is globular. Peptide itself forms an alpha
helix. In Figure 1, the two hydrophobic residues are coloured
red in the picture to the left. They are Leu10 and Leu19.
|
However,
having two hydrophobic anchor residues are where the similarities
to the smMLCK and skMLCK peptides end. First of all, the
spacing between the anchoring residues are down from 12
to 8. Second, the glutamate residues of Ca2+-calmodulin
participates more so than in the smMLCK or skMLCK. Many
side chains in the calmodulin show different conformation
from the smMLCK or skMLCK story. In addition, the linker
region between the two domains show more flexibility than
previously thought by unravelling the alpha-helix further.
Here, we begin to see why calmodulin is able to bind so
many peptides without apparent sequence homology. First,
methionine residues in the binding pocket provide the
variety in the anchoring residues. Second, the linker
region is able to change its conformation to accomodate
many different peptides of different lengths. Third, calmodulin
changes its conformation drastically upon binding calcium
ions. In combination, it is no wonder there are so many
different peptides that bind calmodulin without apparent
sequence homology. |
|
|
|
| |
| |
|
|
| |
|
|